Welcome to the Flavobacterium psychrophilum Expression Data Browser
This website is dedicated to the exploration of the condition-dependent transcriptome of Flavobacterium psychrophilum in different lifestyles.
Université Paris-Saclay, INRAE, VIM and MaIAGE, 78350 Jouy-en-Josas, France
Related article (to use for citation):
Guérin, C., Lee, BH., Fradet, B., van Dijk, E., Mirauta, B., Thermes, C., Bernardet, JF., Repoila, F., Duchaud, E., Nicolas, P. & Rochat, T. Transcriptome architecture and regulation at environmental transitions in flavobacteria: the case of an important fish pathogen. ISME COMMUN.1, 33 (2021). https://doi.org/10.1038/s43705-021-00029-9
This work was financially supported by the French National Research Agency (grant ANR-17-CE20-0020-01 FlavoPatho project).
Summary
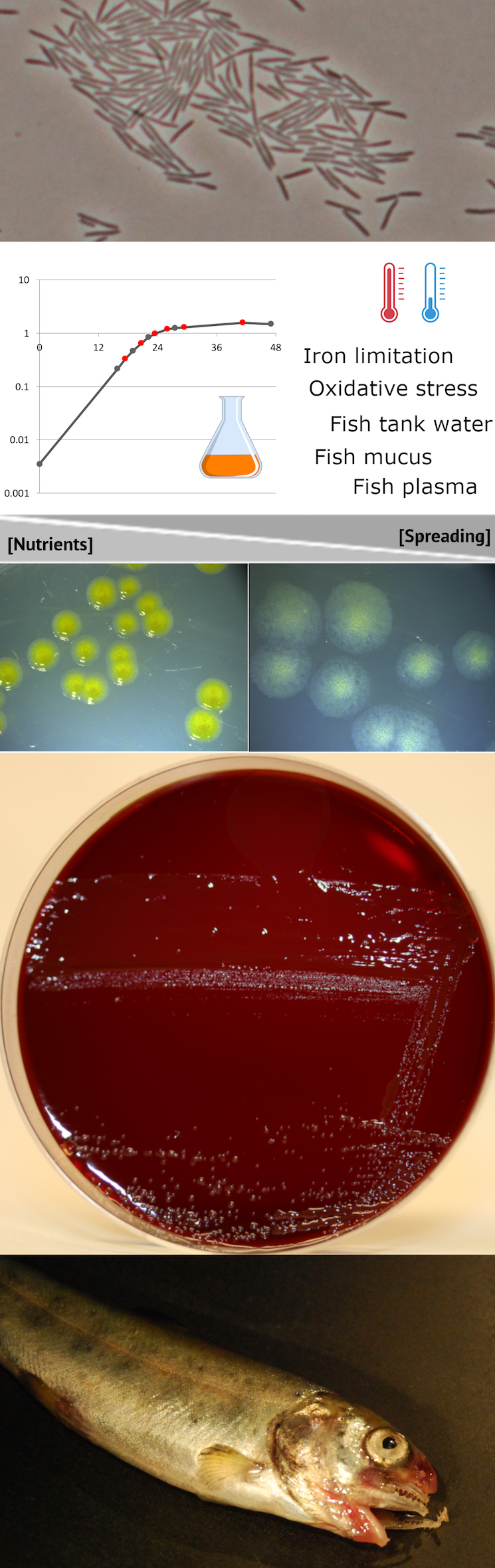
The website fpeb provides a user-friendly access to the data and results. It allows to explore the enriched transcriptional map and experimental data, either along the chromosome or across the expression space (based on correlation between genes expression profiles).
fpeb allows interactive recovery of specific lists of interest (by RNA categories, gene clusters, differential expression in one condition), the export of figures such as expression profile across 32 biological conditions, genomic coordinates and nucleotide sequence for all features (CDS and new RNAs). The study was performed using OSU THCO2-90, a model strain for F. psychrophilum genetic manipulation (Bertolini et al., 1994). In order to facilitate access to specific regions of interest by the entire Flavobacterium research community, the genome browser implements sequence-similarity search.
A selection of tracks to display in the JBrowse view is available, including:
- Structural annotation of all transcribed regions classified into RNA categories
- Transcription start sites with predicted sigma factor promoters
- Read coverage profiles for 5’-end and global RNA-Seq libraries
- Probe-level microarray hybridization signal along the genome of 64 samples
- Probes of the microarray
- Predicted intrinsic terminators
List of differentially expressed genes is computed for 19 pairwise comparisons.